SARS-CoV-2 RT-PCR
Code no.: Pc-7210
Packing contains:
- 20 lyophilized reaction tubes for cDNA synthesis
- 20 lyophilized reaction tubes for E N PCR
- 20 lyophilized reaction tubes for ORF1b-nsp14 PCR
2x 1000 µl PCR-grade water
![]()
![]()
General information:
The SARS-CoV-2 RT-PCR is for in vitro or research use only. This test detects gene sequences of the SARS-CoV-2 genome (NCBI reference: MN908947.3) via Nucleic Acid Amplification Technology (NAAT). It is a 2-step lyophilized RT-PCR. The used primer system is based on the publication of Corman et al. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR from Berlin University Hospital Charité (Drosten group) and Chu et al. Molecular Diagnosis of a Novel Coronavirus (2019-nCoV) Causing an Outbreak of Pneumonia from Hongkong University (Peiris group).
The Cytecs SARS-CoV-2 RT-PCR detects E gene and N gene in a combined assay and ORF1b-nsp14 in a single assay. Test results will be analysed by the means of gel-electrophoresis.
Sensitivity and Specificity:
Detection of beta coronavirus SARS-CoV-2 genes E gene, N gene and ORF1b-nsp14.
Diagnostical sensitivity: 95,45 %
Diagnostical specificity: 98,46 %
Analytical sensitivity:
E N assay: 62.5 copies/ reaction
ORF1b-nsp14 assay: 62.5 copies/ reaction
Analytical sensitivity:
The in silico investigation via BLAST analysis showed no cross reactivity with other species.
An experimental investigation was performed in the publications from Croman et al. and Chu et al.:
HCoV-NL631,2
HCoV-229E1,2
HCoV-OC431,2
HCoV-HKU11,2
MERS-CoV1,2
CCoV-HKU232
Influenza A (H1N1)2
Influenza A (H1N1/09)1
Influenza A (H3N2)2
Influenza A (H5N1)1,2
Influenza A (H7N9)2
Influenza B1
Avian-Influenza (H1)2
Avian-Influenza (H4)2
Avian-Influenza (H6)2
Avian-Influenza (H9)2
Rhinovirus1,2/ Enterovirus1,2
Respiratory syncytial virus (A/ B)1,2
Parainfluenza 1 virus1
Parainfluenza 2 virus1
Parainfluenza 3 virus1,2
Parainfluenza A or B virus1
Human metapneumovirus1,2
Adenovirus1,2
Human Bocavirus1,2
Legionella spp.1
Mycoplasma spp.1
1Corman et al.
2Chu et al.
SARS-CoV-1 does not circulate in the world population – last time in the year 2003 (Wilder-Smith et al., 2020). A cross reactivity with SARS-CoV-1 is nearly impossible.
Procedure:
cDNA synthesis:
The procedure of SARS-CoV-2 RT-PCR starts with unpacking and marking of the cDNA tubes. Rehydrate the pellet with 15 µl of PCR-grade water (included in scope of delivery). Mix gently by pipetting. Incubate the vial for 2 min, ad 5 µl RNA and mix gently.
PCR:
Unpack and mark the PCR-tubes for E N and ORF1b-nsp14 assay and rehydrate the pellet with 23 µl of PCR-grade water (included in scope of delivery). Mix gently by pipetting. Incubate the vials for 2 min, ad 2 µl cDNA in each assay and mix gently.
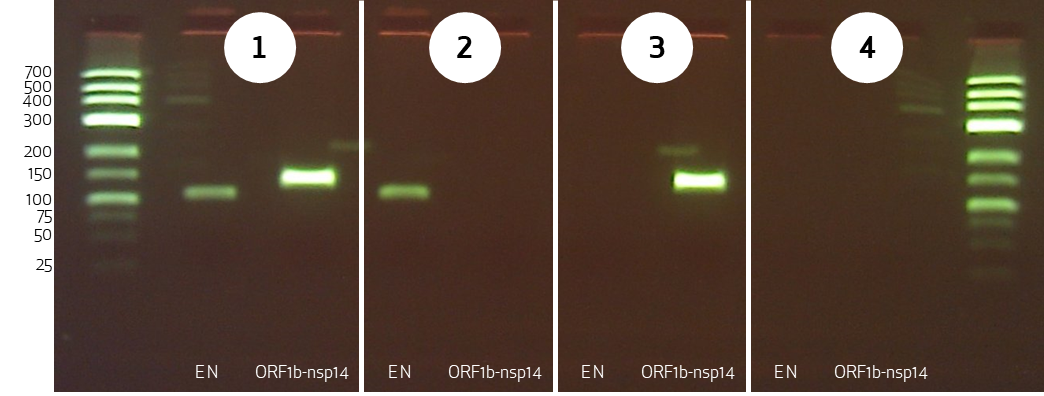
PCR products will be analysed by the means of gel-electrophoresis. The valuation will be described in the next chapter. Figure 2 shows the possible results in an agarose-gel.
Valuation of SARS-CoV-2 RT-PCR test:
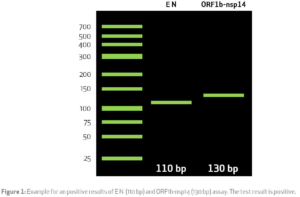
For analysis the PCR products has to be separated in an agarose-gel. The expected result is shown in figure 1.
E N shows a band on the hight of 110 bp and the ORF1b-nsp14 assay shows a band on the hight of 130 bp.
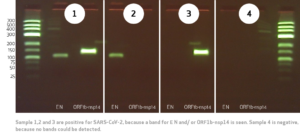
Valuation:
The valuation, if the test is positive or negative will be done corresponding to the band distribution in the agarose-gel. The possible valuations of the SARS-CoV-2 RT-PCR test is shown in figure 2.
Test limitation:
The Cytecs SARS-CoV-2 RT-PCR test belongs to the NAAT tests. This technique could be influenced negatively by the following parameters:
- RNA is instable if not handled with care.
- Specimen collection must be performed well. The SARS-CoV-2 RT-PCR was evaluated only with naso- or oropharyngeal swabs. Other specimens are not allowed as sample material.
- Swabs should be suitable for naso- or oropharyngeal specimen collection.
- Sample must be transferred to the laboratory immediately.
- Extreme conditions during transport like to high temperatures must be excluded.
- Working in accordance with Good Laboratory Practice conformity is necessary.
- The SARS-CoV-2 genome can mutate (Forster et al., 2020). This can occur in primer mismatch.
- E gene exists also in SARS-CoV-1. SARS-CoV-1 does not circulate in the world population. A confusion is not possible.
- The viral load differs during the infection development. False negative results are seen at the beginning and at the end of a SARS-CoV-2 infection. (Kucrika et al., 2020). Test should be repeated if the results are in contrast with the patient’s clinical parameters.
- False positive results may happen from cross contaminations due to bad sample or product handling.
- Possible causes of false negative results:
- Incorrect sample collection
- Extreme transport conditions
- RNA degradation
- Mutation of primer binding site
- RT-PCR inhibitors (e.g. Haemoglobin)
- Failure to follow user manual
NAAT test results should not be the sole basis of decisions in patient management.
Analyzing PCR products with the gel-electrophoresis system E-CUBE
Storage:
The ready to use reaction tubes should be stored at room temperature. Avoid freezing, high temperatures and high humidity.
Literature:
M. Corman, O. Landt, M. Kaiser et al., Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR, Euro surveillance : bulletin Europeen sur les maladies transmissibles = European communicable disease bulletin, vol. 25, no. 3, 2020.
K. W. Chu, Y. Pan, S. M. S. Cheng et al., Molecular Diagnosis of a Novel Coronavirus (2019-nCoV) Causing an Outbreak of Pneumonia, Clinical chemistry, vol. 66, no. 4, pp. 549–555, 2020.
Wilder-Smith A., Chiew C. J., Lee V.J., Can we contain the COVID-19 outbreak with the same measures as for SARS?, Lancet Infect Dis 2020; 20: e102–07, doi: https://doi.org/10.1016/ S1473-3099(20)30129-8.
Forster, L. Forster, C. Renfrew, et al., Phylogenetic network analysis of SARS-CoV-2 genomes, Proceedings of the National Academy of Sciences of the United States of America, vol. 117, no. 17, pp 9241-9243, 2020.
L. M. Kucirka, S. A. Lauer,O. Laeyendecker et al., Variation in False-Negative Rate of Reverse Transcriptase Polymerase Chain Reaction-Based SARS-CoV-2 Tests by Time Since Exposure, Annals of internal medicine, DOI: 10.7326/M20-1495, 2020.